INSTRUCTIONS
■ Tracks
-
1. Reference sequence
- Reference sequence: Human genome hg19/GRCh37 sequence.
-
2. IMM: 3 cell-types
- CpG_CD4T, CpG_Mono, CpG_Neu: Information of each CpG site.
- CpG_CD4T_avg, CpG_Mono_avg, CpG_Neu_avg: Average methylation levels of each CpG site.
- CpG_CD4T_sd, CpG_Mono_sd, CpG_Neu_sd: SD (standard deviations) for the methylation levels of each CpG site.
- CpG_CD4T_RI, CpG_Mono_RI, CpG_Neu_RI: RI (reference interval) for the methylation levels of each CpG site.
- FPKM_CD4T, FPKM_Mono, FPKM_Neu: FPKM (fragments per kilobase of exon per million fragments mapped) values of each transcript.
- SNV_CD4T, SNV_Mono, SNV_Neu: SNV (single nucleotide variant, minor allele count > 1 and call rate ≥ 50%).
-
3. IMM: 8 cell-populations
- All tracks: Average methylation levels of each CpG site of each cell-population obtained from WGBS of 20 individuals.
-
4. IMM: QTL
- eQTL: Results of eQTL analysis (only the results with FDR ≤ 0.05 are presented).
- eQTM: Results of eQTM analysis (only the results with FDR ≤ 0.05 are presented).
- mQTL: Results of mQTL analysis (only the results with FDR ≤ 0.05 are presented).
-
5. Others
- RepeatMasker: Repetitive elements from UCSC genome browser.
- CpGIslandsExt: CpG Island locations obtained from UCSC genome browser.
- HM450: probe information of Illumina Infinium HumanMethylation450 Array BeadChips.
- genecode_v19: Information of genes obtained from GENCODE version 19.
- genecode_v19_trs: Information of transcripts obtained from GENCODE version 19.
■ How to Use
Click “Genome Browser” tab in the navigation menu, and search a gene or region of your interest by following the instructions below.
■ Search methods
- Search by Gene Symbol
- Enter any gene symbol in the box and press the Go button.

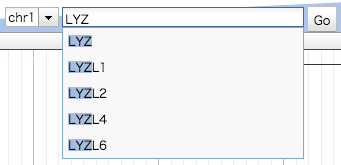
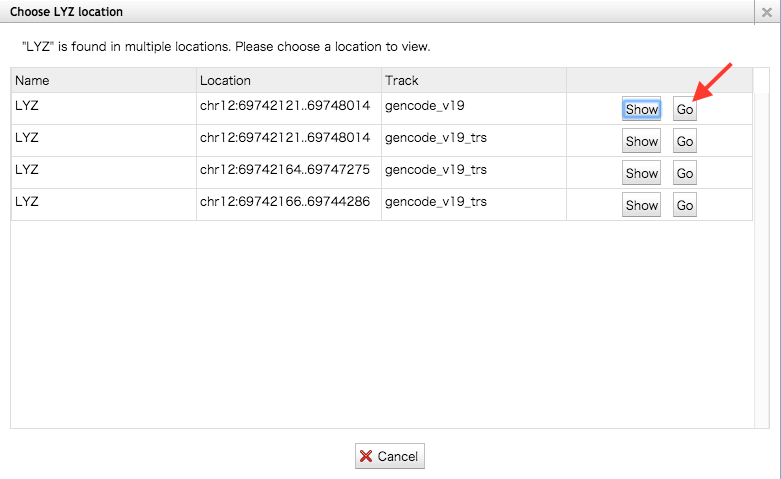
- Ex; Enter the “LYZ” and press the button. Choose gencode_v19 from the pop-up menu and press the Go button.
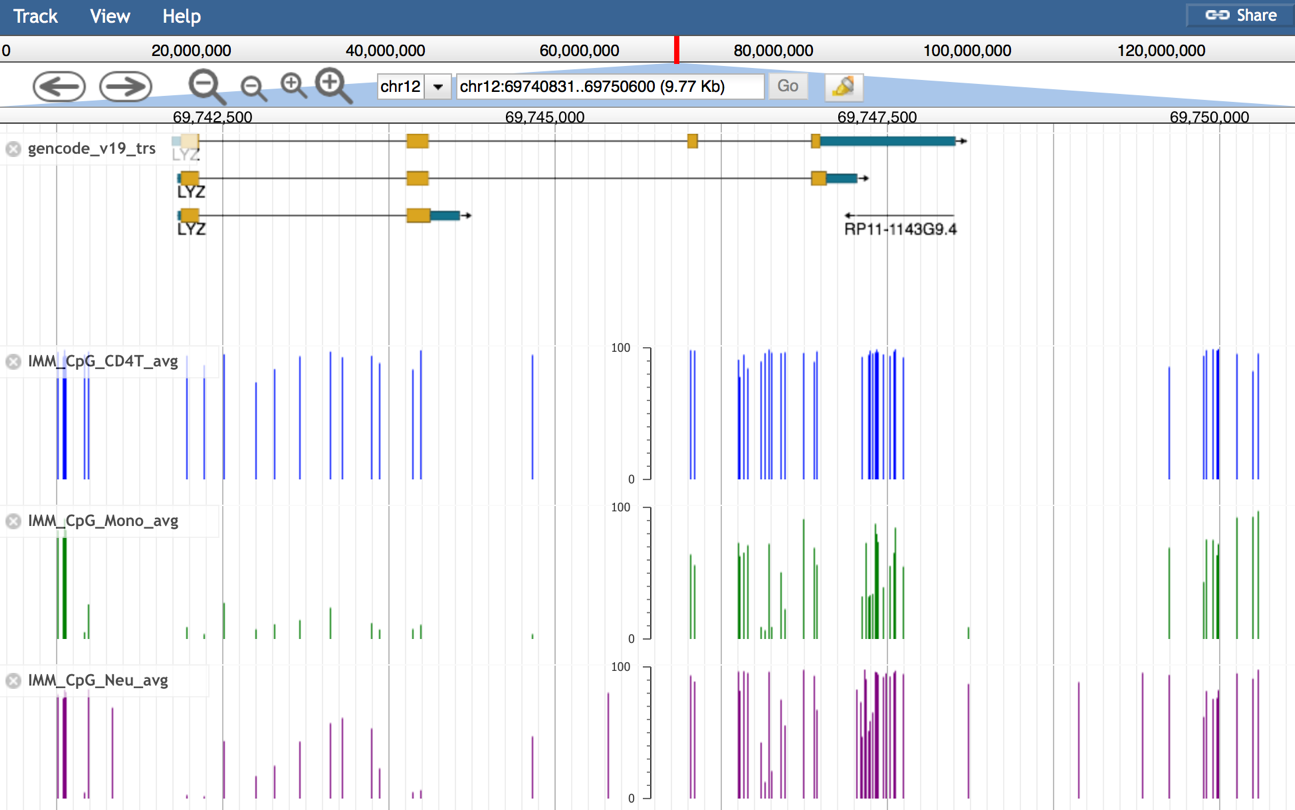
- You can find differencially methylated CpG sites among cell types at the LYZ gene region.
- Search by Genomic Region
- Enter any genomic position or region in the box and press the Go button.
- Ex; Select “chr1” from select box and enter the “206906382” in the box, and then press the Go button.

- You can find CD4+ T cells specific demethylation CpG site at the intergenic region between IL10 and IL19.
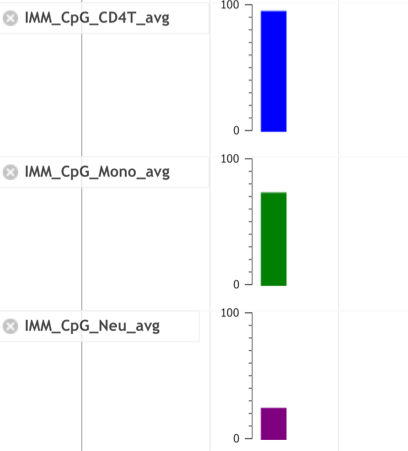
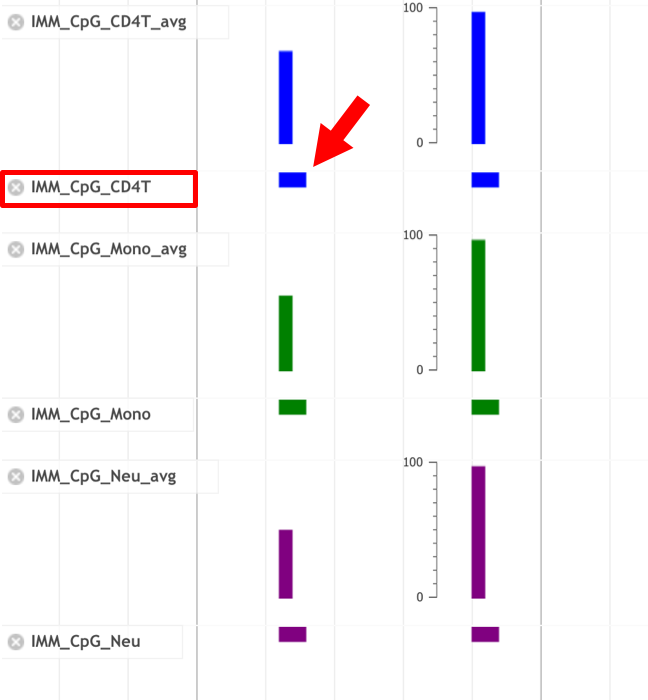
■ Details of CpG track
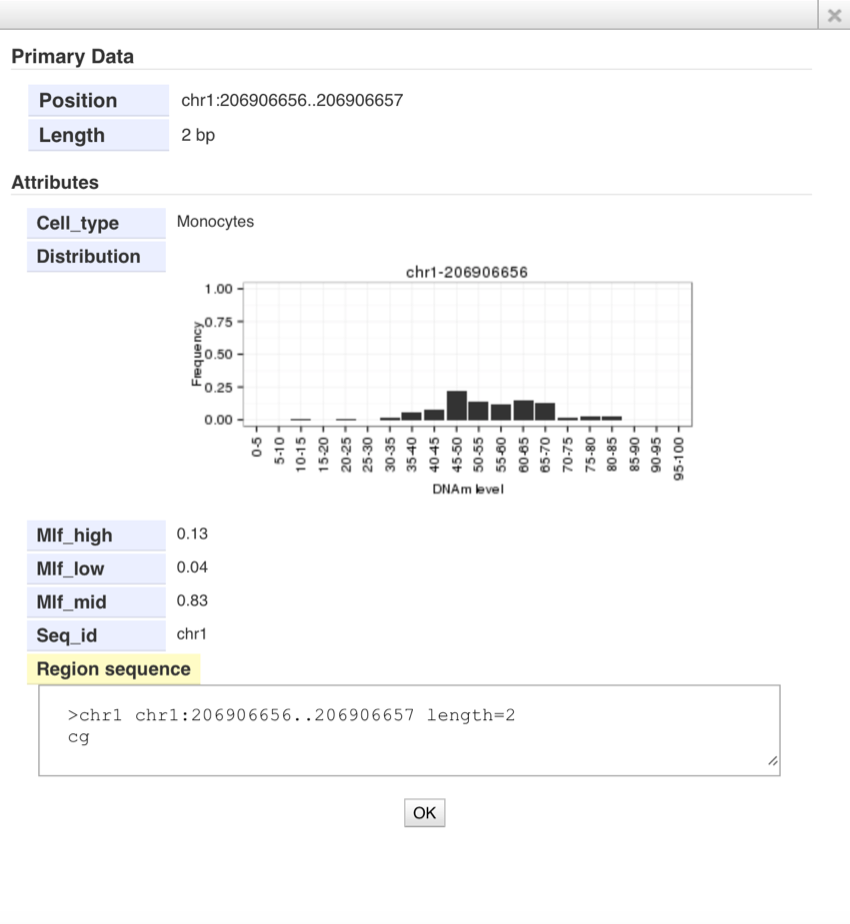
- Click the bar in the CpG tracks. You can see distribution of methylation levels of each CpG site in the pop-up window.
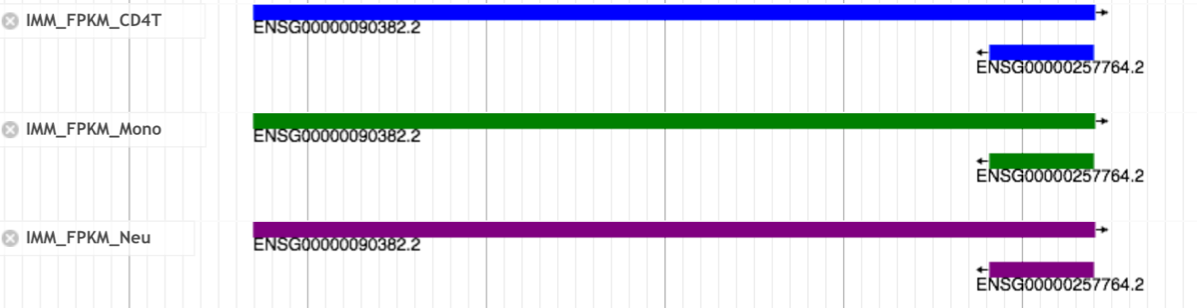
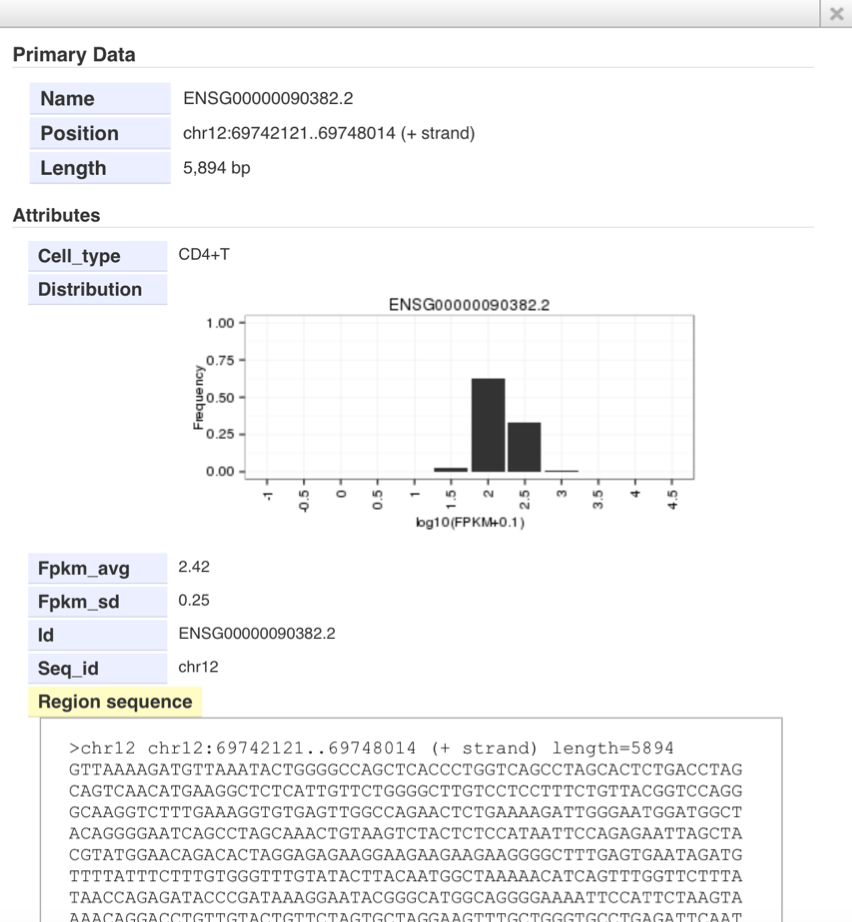
- Details of the FPKM track
- Click the bar in the FPKM tracks. You can see distribution of expression levels of each gene in the pop-up window
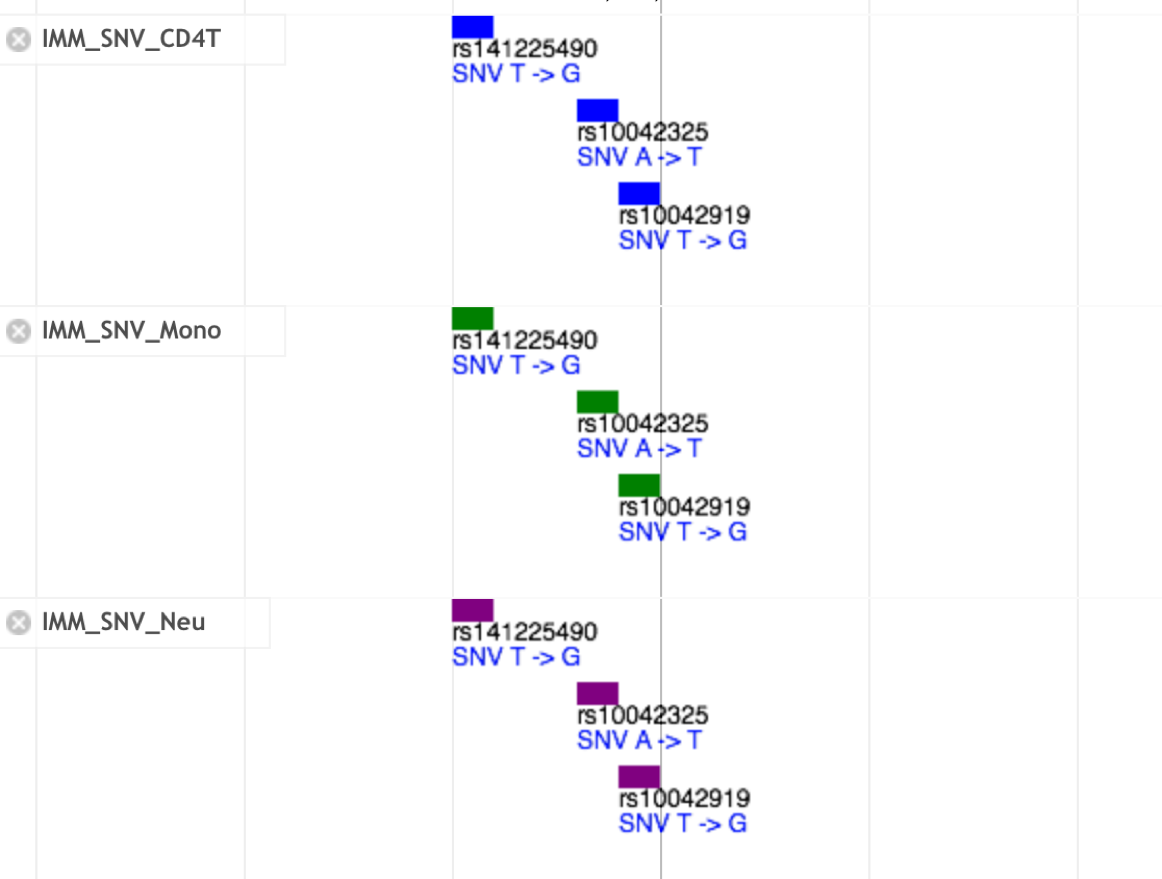
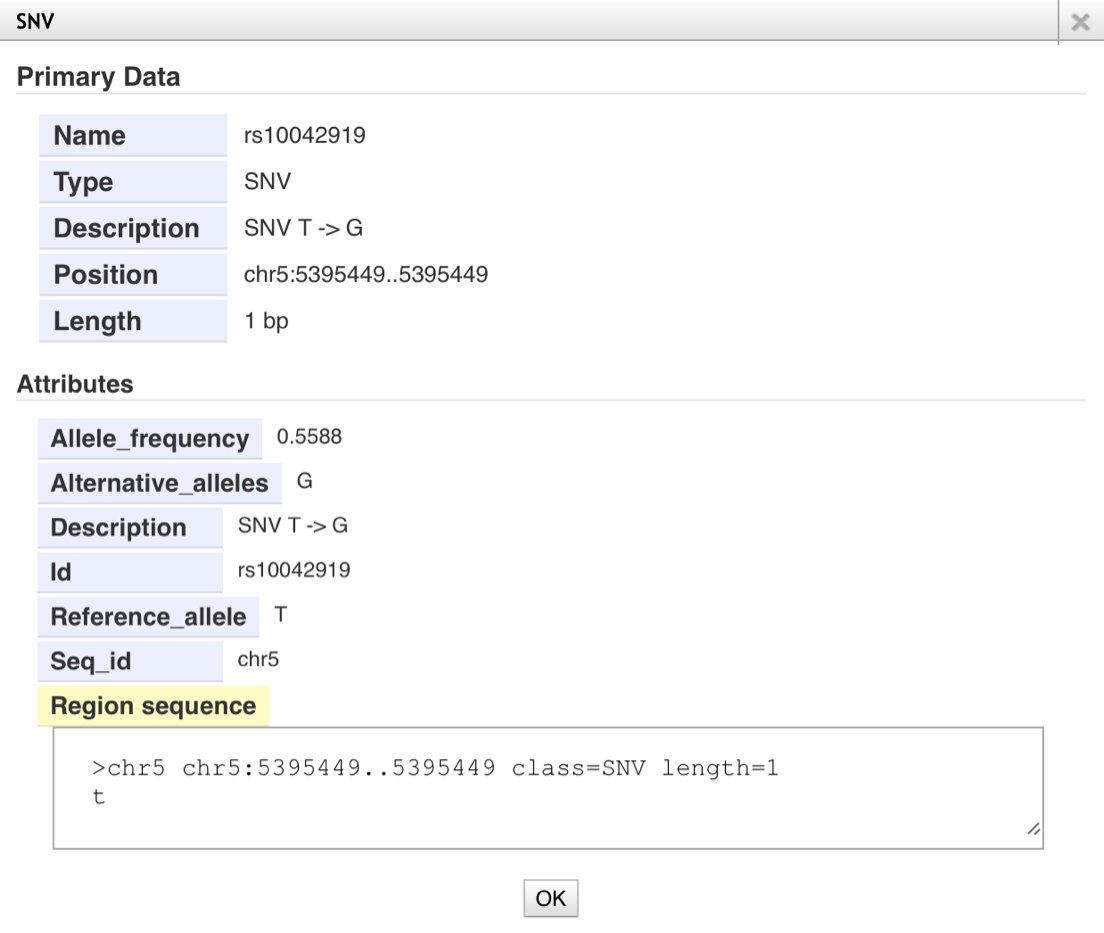
- Details of the SNV track
- Click the bar in the SNV track. You can see detail information of each SNV in the pop-up window.
- Terms
- Cell Type; Name of each cell type (Mono; monocytes, CD4T; CD4+ T cells, Neu; neutrophils)
- Mlf_high,Mlf_mid, and Mlf_low; Frequency of 102 individuals in each category.
Mlf_high means more than 66% methylation, Mlf_mid means between 34-66%
methylation, and Mlf_low means less than 34% CpG methylation.
- Allele_frequency; Proportion of alternative allele among the total alleles for the subjects in the SNV tracks.